Flaws are present in every tool for editing every file type, and despite the fact that you can use many solutions out there, not all of them will fit your specific needs. DocHub makes it easier than ever to make and alter, and handle paperwork - and not just in PDF format.
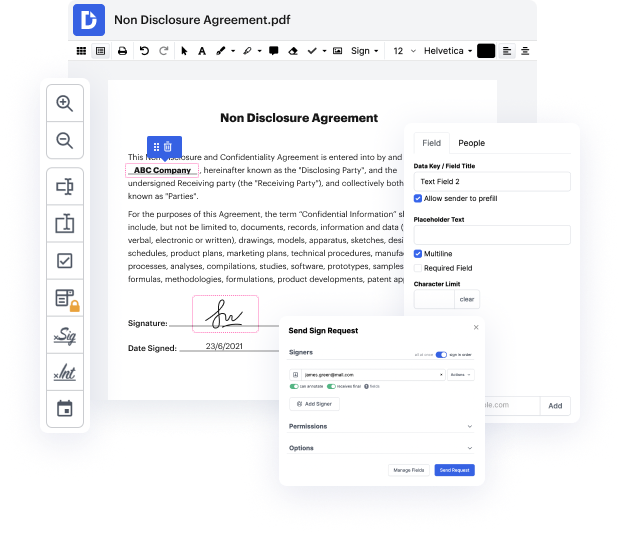
Every time you need to easily inlay questionaire in FTM, DocHub has got you covered. You can easily modify form components including text and images, and structure. Customize, arrange, and encrypt files, build eSignature workflows, make fillable forms for stress-free information gathering, etc. Our templates feature allows you to create templates based on paperwork with which you often work.
In addition, you can stay connected to your go-to productivity tools and CRM solutions while handling your files.
One of the most incredible things about utilizing DocHub is the ability to manage form activities of any difficulty, regardless of whether you require a quick modify or more complex editing. It comes with an all-in-one form editor, website form builder, and workflow-centered tools. In addition, you can be certain that your paperwork will be legally binding and adhere to all security protocols.
Shave some time off your projects with DocHub's capabilities that make handling files effortless.
This video uses three examples from the chicken genome to illustrate common uses of clone placement data: - Find genomic clones that contain a gene of interest - Use clone placements to recognize a genomic region with potential mis-assembly - Use clone placements to recognize potential structural variations A companion video that focuses on clone FTP content is linked at the end of this video. Very briefly, clone placements are created by aligning end and insert sequences on genome assemblies annotated at NCBI. To generate a clone placement for end sequences, both ends (forward and reverse) must be placed on the same assembly molecule. For the first example, letamp;#39;s find clone placements in the region of the chicken WDR3 gene. Iamp;#39;m starting in the Genome Data viewer, but if you happened to be in a Gene record, you could link from there to the same GDV display we are about to see. Iamp;#39;ll select the previous chicken assembly, Gallus-gallus 5.0, but that is simply becau