Not all formats, such as docbook, are created to be easily edited. Even though numerous features can help us change all document formats, no one has yet invented an actual all-size-fits-all tool.
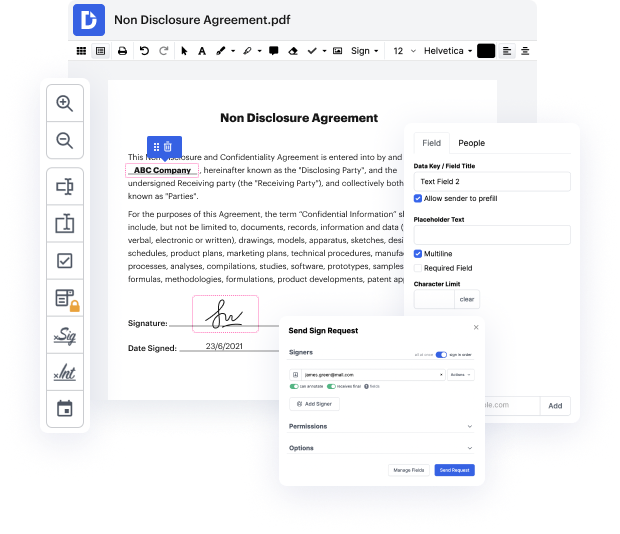
DocHub gives a easy and streamlined tool for editing, managing, and storing papers in the most widely used formats. You don't have to be a tech-knowledgeable person to adjust epitaph in docbook or make other modifications. DocHub is robust enough to make the process straightforward for everyone.
Our tool allows you to modify and tweak papers, send data back and forth, generate dynamic forms for information collection, encrypt and safeguard documents, and set up eSignature workflows. Additionally, you can also generate templates from papers you utilize regularly.
You’ll find plenty of other functionality inside DocHub, such as integrations that let you link your docbook document to various business programs.
DocHub is an intuitive, fairly priced way to deal with papers and streamline workflows. It offers a wide selection of features, from generation to editing, eSignature services, and web document creating. The software can export your files in multiple formats while maintaining highest protection and following the maximum information safety requirements.
Give DocHub a go and see just how straightforward your editing process can be.
so while this is loading yes my name is thiago palma i will be talking about a map which is protein family database which you can use to determine if a protein belong to the family and what the function is and now weamp;#39;re talking about how we extend that binary match and to actually annotating them because so the problem is we have very few annotated proteins and many many more which are not annotated and everywhere so the problem is we got a family of proteins and if we know which families they belong to then we can annotate them i can do this with rules for example if it matches hem up ms j 1 to 1 then it has this EC number and this is standard biology this is pretty good it increases you know our coverage from 1 in 200 20 to 25% to 64 percent and we can actually express these rules as sparkle queries so you have the protein identification is done with InterPro scan the summary tools to translate that into rdf and they havenamp;#39;t that arenamp;#39;t yes we can then use thi